BMRB Featured System: Ubiquitin |
|||||
| |||||
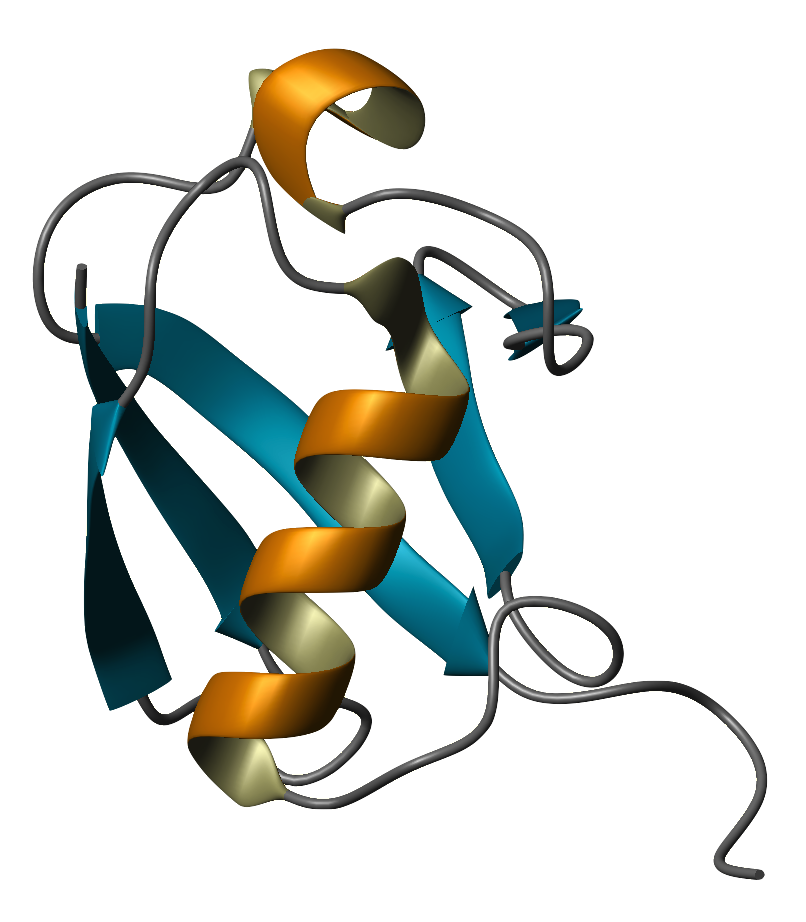
Although completely unknown a little more than 30 years ago, ubiquitin (Ub) now counts among the most intensely studied proteins. The 2004 Nobel Prize in Chemistry was awarded to the researchers who elucidated its most prominent function - tagging proteins for ATP dependent degradation. Ubiquitin is a small protein that does not have any one function, but acts as a mediator for a number of other processes. Ubiquitin is found only in eukaryotic organisms, but it is found in all of them. Its ubiquity within the eukaryotes is what gives it its name. Evolutionarily, the sequence of ubiquitin is highly conserved, with a difference of only a few residues between plants, yeasts, and animals. It has a characteristic geometry, called the Ub-fold, which consists of beta-strands that flank an alpha helical core. This is a common fold in nature, that appears as a domain embedded in larger, more complex structures. The fold is also found in ubiquitin-like proteins that may have a low sequence identity with Ub, but appear to be evolutionarily related. Thus although Ub is found only in eukaryotes, Ub-like proteins are found also in archaea and eubacteria. Ubiquitin was discovered by Gideon Goldstein and colleagues in the mid-1970s while they were studying an autoimmune, neuromuscular disease called myasthenia gravis. It was originally isolated along with several other proteins, from bovine thymus and thought to be a thymic hormone. By 1975, it had been found to be very widespread in living cells and was given the name, ubiquitous immunopoietic polypeptide (UBIP), which was soon shortened to ubiquitin. i ii In 1979 and 1980 Avram Hershko, Aharon Ciechanover, and Irwin A. Rose proposed a mechanism for ATP dependent breakdown of proteins. iii iv As it turned out, ubiquitin was a key element of this system. It was this work that led to their receiving the Nobel Prize in Chemistry in 2004. Since then, Ub has been found to play a role in other functions such as silencing the redundant X chromosome in female mammals and membrane trafficking. Structure-function studies of Ub and Ub-like proteins form an active and dynamic field of inquiry. Ubiquitin itself consists of 76 amino acid residues (molecular mass 8500), although the early literature often describes it as having 74 residues, because the two C-terminal glycine residues were often cleaved off during preparation. The protein is stable over a wide range of temperature and pH. Its C-terminal residues form an unencumbered coil ending in an arginine followed by two glycine residues. As you will recall, glycine has no R-group other than a hydrogen atom, which leaves the terminating carboxylic acid free to bond to other proteins, forming an amide with a lysine residue. Weak thioester bonds will be formed between the terminal glycine and cysteine residues of Ub-conjugating and Ub-ligating enzymes during the process of ubiquitination, but these are only transitive. Ub contains seven lysine residues of its own, each of which is able to bond to another Ub, forming a poly-ubiquitin chain. It appears that the linkage positions and number of linkages determines the fate of the tagged protein. Tacking a Ub onto another protein is a process called either ubiquitination (or ubiquitinylation). Some people feel that the process of tagging another molecule with Ub is analogous to phosphorylation and thus that ubiquitinylation is the more appropriate term. The Protein Data Bank and the European Bioinformatics Institute both chose Ubiquitin as their Protein of the Month for December of 2004. These sites have additional information and structural details. Richard Haris at the UCL/LICR Joint NMR Laboratory maintains an NMR resource page for ubiquitin. | |||||
|
Next: Ub's function |